4. Clustering
[1]:
getData <- function(N=30) {
x1 <- c(
rnorm(N, mean=2.5, sd=2.5),
rnorm(N, mean=10.5, sd=2.5)
)
x2 <- c(
rnorm(N, mean=2.5, sd=2.5),
rnorm(N, mean=10.5, sd=2.5)
)
y <- c(rep(0, N), rep(1, N))
X <- data.frame(x1=x1, x2=x2)
return(list(X=X, y=y))
}
D = getData()
4.1. K-means
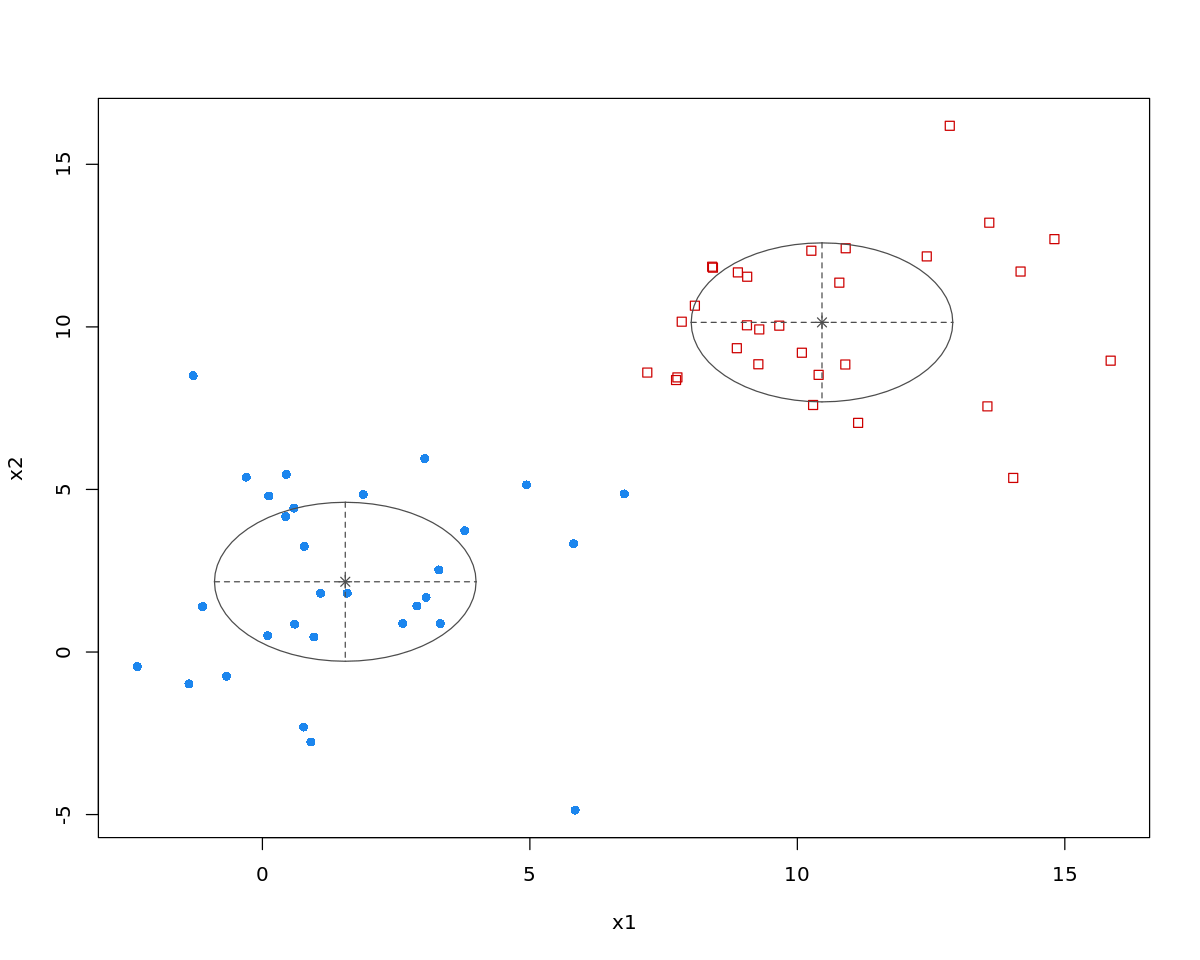
[2]:
m <- kmeans(D$X, 2)
[3]:
print(m)
K-means clustering with 2 clusters of sizes 30, 30
Cluster means:
x1 x2
1 10.519288 10.215782
2 1.618027 2.197318
Clustering vector:
[1] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 1 1 1 1 1 1 1
[39] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
Within cluster sum of squares by cluster:
[1] 306.8271 403.1129
(between_SS / total_SS = 75.2 %)
Available components:
[1] "cluster" "centers" "totss" "withinss" "tot.withinss"
[6] "betweenss" "size" "iter" "ifault"
[4]:
print(m$totss)
[1] 2862.863
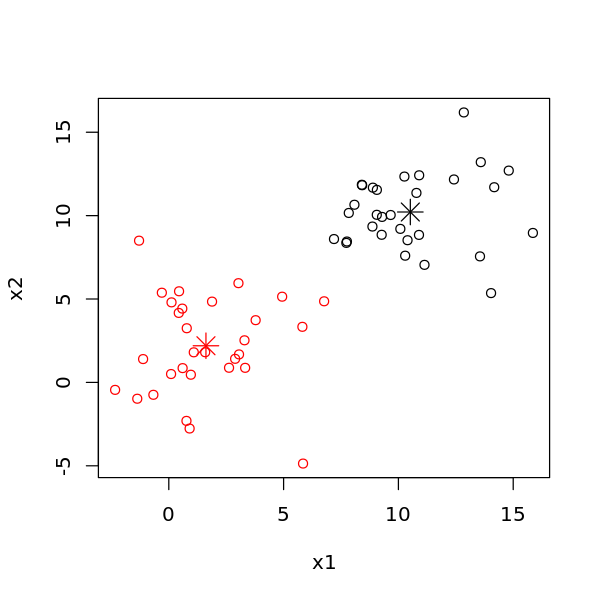
[5]:
options(repr.plot.width=5, repr.plot.height=5)
plot(D$X, col=m$cluster)
points(m$centers, col=1:2, pch=8, cex=2)
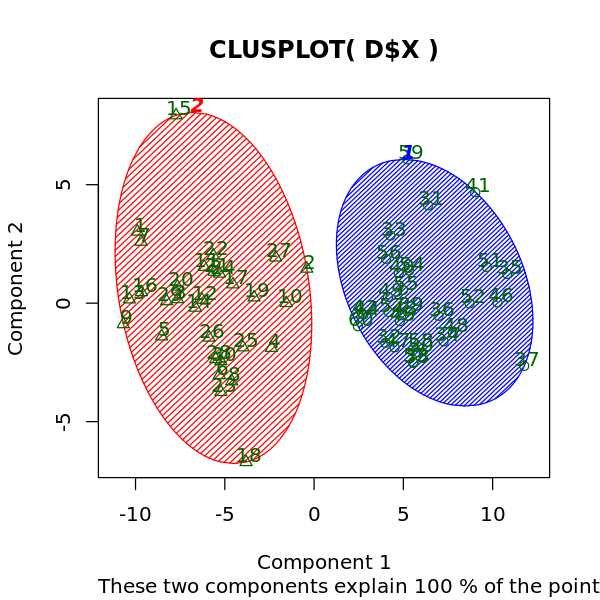
[6]:
library('cluster')
options(repr.plot.width=5, repr.plot.height=5)
clusplot(D$X, m$cluster, color=TRUE, shade=TRUE, labels=2, lines=0)
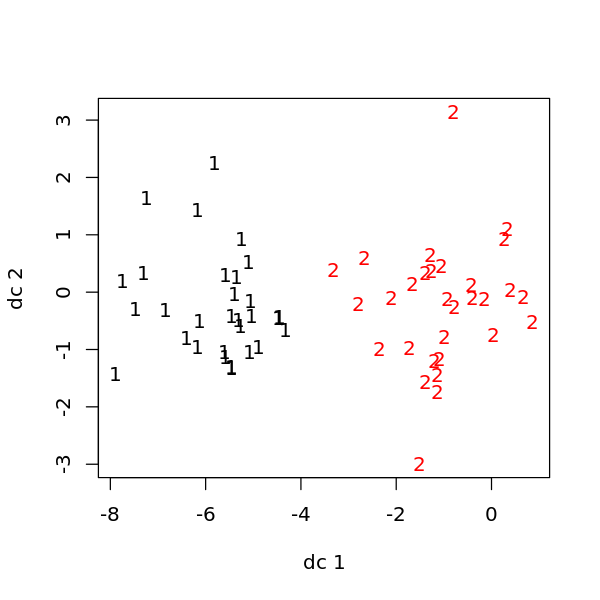
[7]:
library('fpc')
options(repr.plot.width=5, repr.plot.height=5)
plotcluster(D$X, m$cluster)
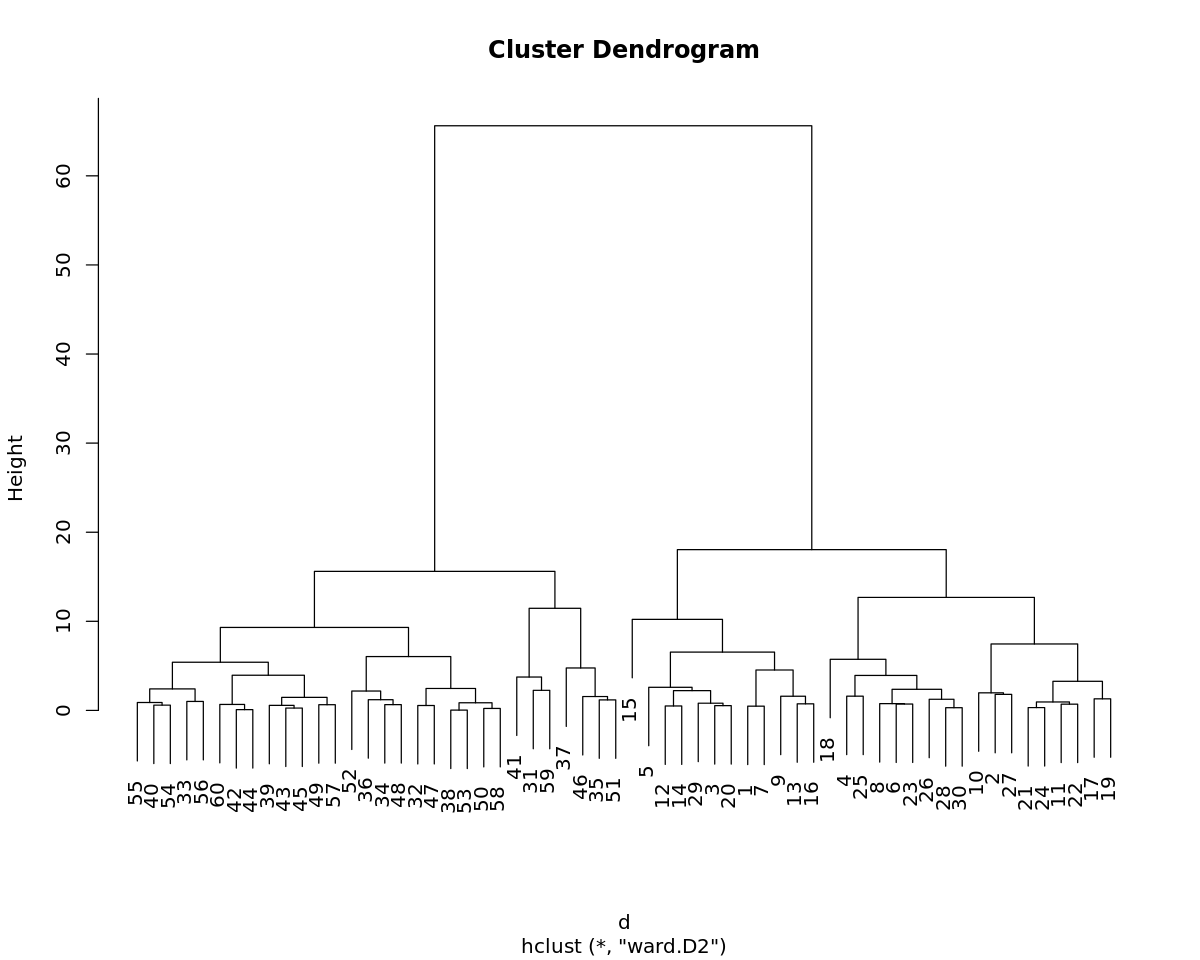
4.2. Ward hierarchical clustering
[8]:
d <- dist(D$X, method='euclidean')
m <- hclust(d, method='ward.D2')
[9]:
print(m)
Call:
hclust(d = d, method = "ward.D2")
Cluster method : ward.D2
Distance : euclidean
Number of objects: 60
[10]:
options(repr.plot.width=10, repr.plot.height=8)
plot(m)
4.3. Model based
[11]:
library('mclust')
m <- Mclust(D$X)
Package 'mclust' version 5.4.5
Type 'citation("mclust")' for citing this R package in publications.
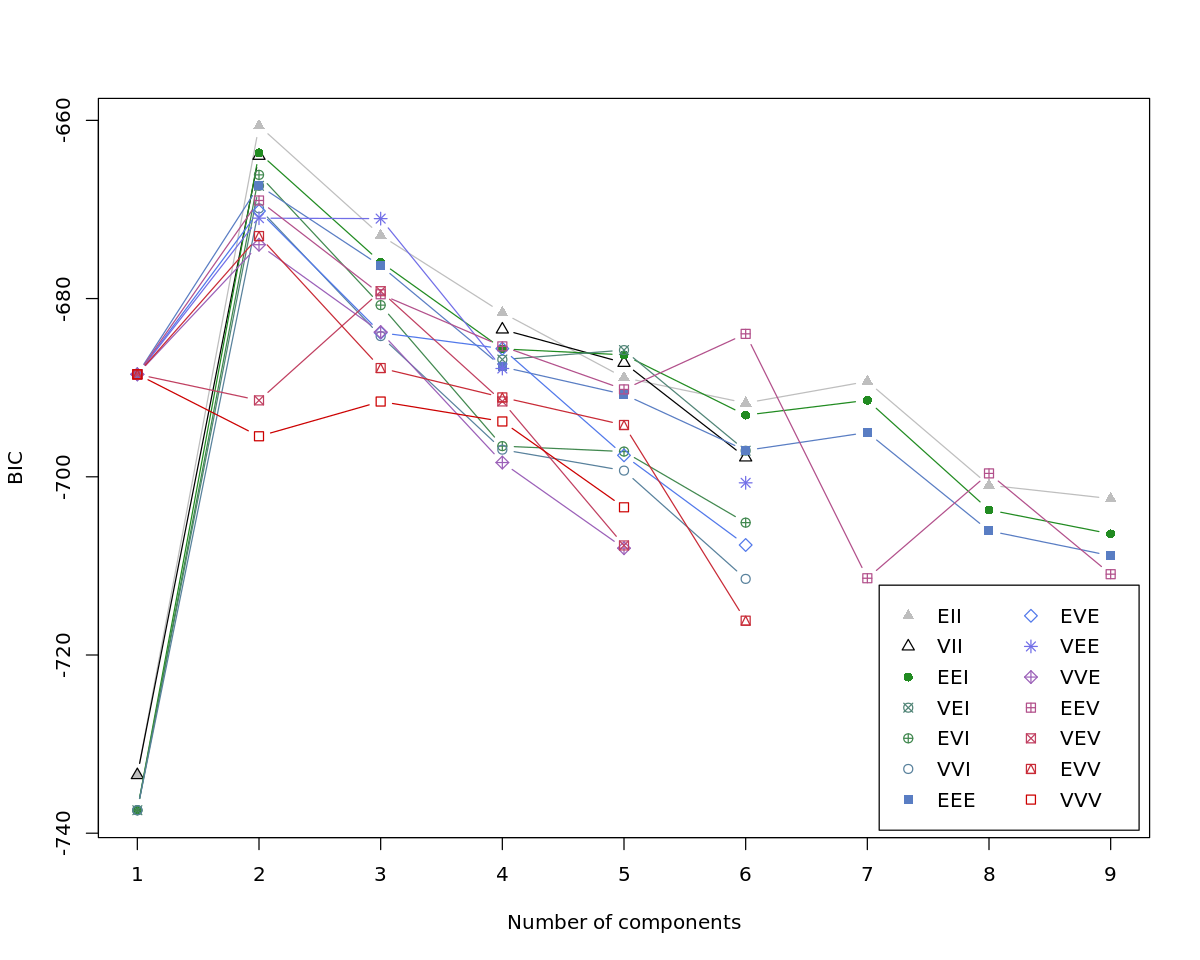
[12]:
print(summary(m))
----------------------------------------------------
Gaussian finite mixture model fitted by EM algorithm
----------------------------------------------------
Mclust EII (spherical, equal volume) model with 2 components:
log-likelihood n df BIC ICL
-318.0198 60 6 -660.6057 -661.7037
Clustering table:
1 2
30 30
[13]:
plot(m)
4.4. Comparing clustering solutions
[14]:
m1 <- kmeans(D$X, 2)
m2 <- Mclust(D$X)
d <- dist(D$X, method='euclidean')
s <- cluster.stats(d, m1$cluster, m2$cluster)
print(s)
$n
[1] 60
$cluster.number
[1] 2
$cluster.size
[1] 30 30
$min.cluster.size
[1] 30
$noisen
[1] 0
$diameter
[1] 10.89581 15.15251
$average.distance
[1] 4.034249 4.647682
$median.distance
[1] 3.649638 4.319535
$separation
[1] 3.633523 3.633523
$average.toother
[1] 12.4389 12.4389
$separation.matrix
[,1] [,2]
[1,] 0.000000 3.633523
[2,] 3.633523 0.000000
$ave.between.matrix
[,1] [,2]
[1,] 0.0000 12.4389
[2,] 12.4389 0.0000
$average.between
[1] 12.4389
$average.within
[1] 4.340965
$n.between
[1] 900
$n.within
[1] 870
$max.diameter
[1] 15.15251
$min.separation
[1] 3.633523
$within.cluster.ss
[1] 709.94
$clus.avg.silwidths
1 2
0.6738004 0.6067246
$avg.silwidth
[1] 0.6402625
$g2
NULL
$g3
NULL
$pearsongamma
[1] 0.8017156
$dunn
[1] 0.2397967
$dunn2
[1] 2.676367
$entropy
[1] 0.6931472
$wb.ratio
[1] 0.348983
$ch
[1] 175.8875
$cwidegap
[1] 3.071360 5.367805
$widestgap
[1] 5.367805
$sindex
[1] 3.88928
$corrected.rand
NULL
$vi
NULL
4.5. Silhouette score
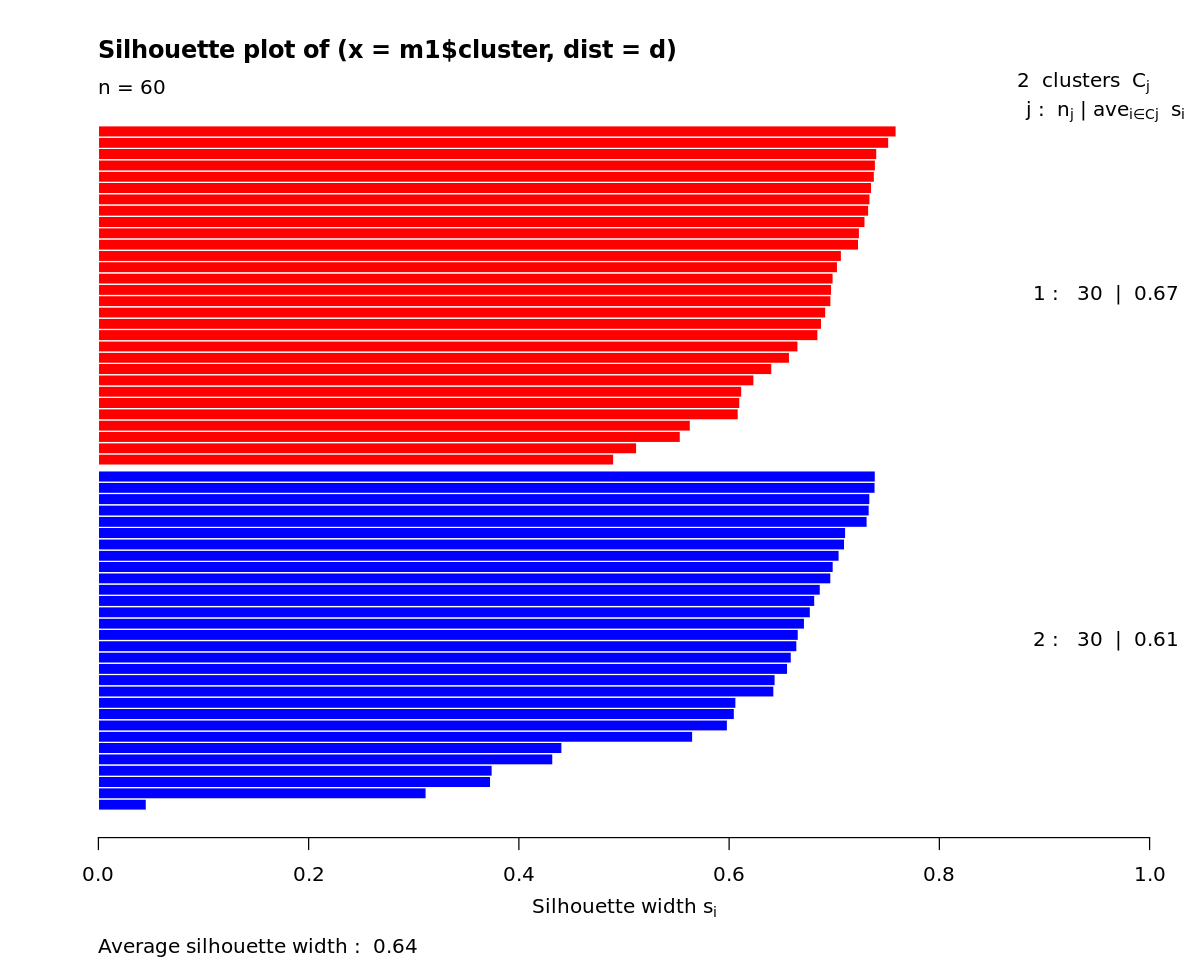
[15]:
s <- silhouette(m1$cluster, d)
print(summary(s))
Silhouette of 60 units in 2 clusters from silhouette.default(x = m1$cluster, dist = d) :
Cluster sizes and average silhouette widths:
30 30
0.6738004 0.6067246
Individual silhouette widths:
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.04566 0.60988 0.68303 0.64026 0.72343 0.75897
[16]:
plot(s, col=c('red', 'blue'))