3. Classification
3.1. Simulate data
Below, we simulate data.
\(X_1 \sim \mathcal{N}(0, 1)\)
\(X_2 \sim \mathcal{N}(0, 1)\)
\(Y_0 = 1 + 2X_1 + 3X_2 + \sigma\)
\(Y_1 = \frac{1}{1 + \exp(-Y_0)}\)
\(y \sim \mathcal{Binom}(Y_1)\)
[1]:
suppressMessages({
library('dplyr')
})
set.seed(37)
getData <- function(N=1000) {
x1 <- rnorm(N, mean=0, sd=1)
x2 <- rnorm(N, mean=0, sd=1)
y <- 1 + 2.0 * x1 + 3.0 * x2 + rnorm(N, mean=0, sd=1)
y <- 1.0 / (1.0 + exp(-y))
y <- rbinom(n=N, size=1, prob=y)
df <- data.frame(x1=x1, x2=x2, y=y)
df <- df %>%
mutate(y=ifelse(y == 0, 'neg', 'pos')) %>%
mutate_if(is.character, as.factor)
return(df)
}
T <- getData()
V <- getData()
print(summary(T))
x1 x2 y
Min. :-2.8613 Min. :-3.28763 neg:394
1st Qu.:-0.6961 1st Qu.:-0.59550 pos:606
Median :-0.0339 Median : 0.06348
Mean :-0.0184 Mean : 0.03492
3rd Qu.: 0.6836 3rd Qu.: 0.69935
Max. : 3.8147 Max. : 3.17901
3.2. Classification methods
There are many classifcation models available in R. We only show a few here and use the pROC package to visualize the performance.
3.2.1. Random forest
Using random forest.
[2]:
suppressMessages({
library('pROC')
library('randomForest')
})
m <- randomForest(y ~ ., data=T, mtry=2, ntree=1000)
[3]:
print(m)
Call:
randomForest(formula = y ~ ., data = T, mtry = 2, ntree = 1000)
Type of random forest: classification
Number of trees: 1000
No. of variables tried at each split: 2
OOB estimate of error rate: 20.6%
Confusion matrix:
neg pos class.error
neg 287 107 0.2715736
pos 99 507 0.1633663
[4]:
p <- predict(m, newdata=V, type='prob')
y_pred = p[,2]
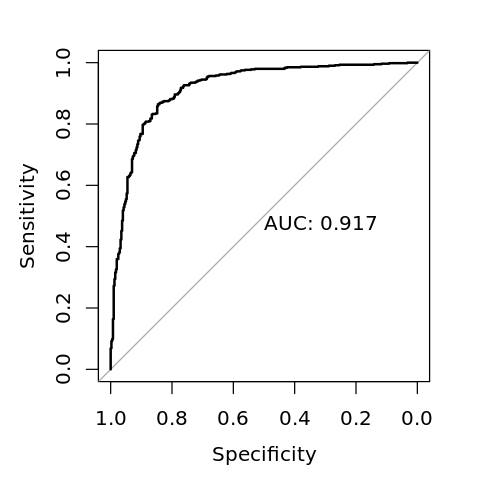
r <- roc(V$y, y_pred, levels=c('neg', 'pos'), direction='<')
print(r)
Call:
roc.default(response = V$y, predictor = y_pred, levels = c("neg", "pos"), direction = "<")
Data: y_pred in 402 controls (V$y neg) < 598 cases (V$y pos).
Area under the curve: 0.9062
[5]:
options(repr.plot.width=4, repr.plot.height=4)
plot(r, print.auc=TRUE)
3.2.2. Logistic regression
Using logistic regression.
[6]:
m <- glm(y ~ ., family='binomial', data=T)
[7]:
print(m)
Call: glm(formula = y ~ ., family = "binomial", data = T)
Coefficients:
(Intercept) x1 x2
0.7692 1.6568 2.3364
Degrees of Freedom: 999 Total (i.e. Null); 997 Residual
Null Deviance: 1341
Residual Deviance: 724.4 AIC: 730.4
[8]:
y_pred <- predict(m, newdata=V, type='response')
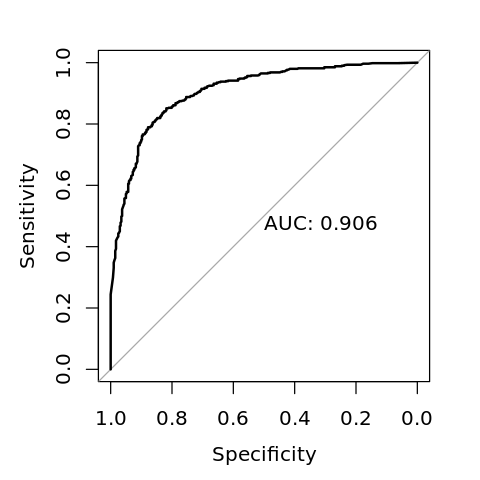
r <- roc(V$y, y_pred, levels=c('neg', 'pos'), direction='<')
print(r)
Call:
roc.default(response = V$y, predictor = y_pred, levels = c("neg", "pos"), direction = "<")
Data: y_pred in 402 controls (V$y neg) < 598 cases (V$y pos).
Area under the curve: 0.933
[9]:
options(repr.plot.width=4, repr.plot.height=4)
plot(r, print.auc=TRUE)
3.2.3. AdaBoost
Using AdaBoost.
[10]:
library('fastAdaboost')
m <- adaboost(y ~ ., nIter=50, data=T)
[11]:
print(m)
adaboost(formula = y ~ ., data = T, nIter = 50)
y ~ .
Dependent Variable: y
No of trees:50
The weights of the trees are:0.96387420.80235240.71526760.63439580.63139020.58530950.62330850.58756820.53374960.5300160.50916270.55428910.54749730.60577360.55118370.54656730.49836120.53329020.48512470.46704230.51634080.52241440.53706090.55988470.55272350.56468040.54326980.53403390.5140650.51328940.54461680.49396760.4945920.45737710.47303330.63182430.52913920.57678280.54805410.53541630.56575640.53455380.53633590.51994930.53944080.56411030.51954350.54601860.48144430.5298128
[12]:
p <- predict(m, newdata=V)
y_pred <- p$prob[,2]
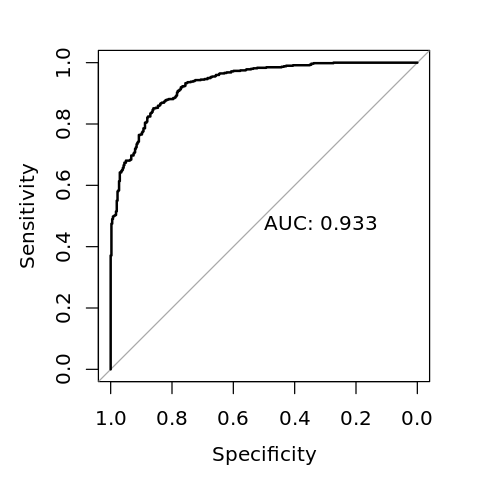
r <- roc(V$y, y_pred, levels=c('neg', 'pos'), direction='<')
print(r)
Call:
roc.default(response = V$y, predictor = y_pred, levels = c("neg", "pos"), direction = "<")
Data: y_pred in 402 controls (V$y neg) < 598 cases (V$y pos).
Area under the curve: 0.8936
[13]:
options(repr.plot.width=4, repr.plot.height=4)
plot(r, print.auc=TRUE)
3.2.4. Support vector machine
Using support vector machine SVM.
[14]:
library('e1071')
m <- svm(y ~ ., probability=TRUE, data=T)
[15]:
print(m)
Call:
svm(formula = y ~ ., data = T, probability = TRUE)
Parameters:
SVM-Type: C-classification
SVM-Kernel: radial
cost: 1
Number of Support Vectors: 405
[16]:
p <- predict(m, newdata=V, probability=TRUE)
y_pred <- attr(p, 'probabilities')[,1]
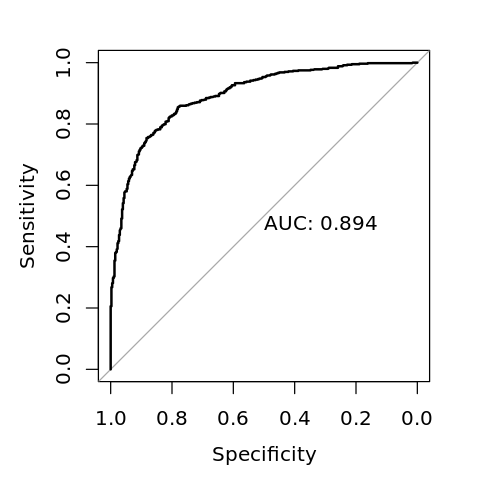
r <- roc(V$y, y_pred, levels=c('neg', 'pos'), direction='<')
print(r)
Call:
roc.default(response = V$y, predictor = y_pred, levels = c("neg", "pos"), direction = "<")
Data: y_pred in 402 controls (V$y neg) < 598 cases (V$y pos).
Area under the curve: 0.9166
[17]:
options(repr.plot.width=4, repr.plot.height=4)
plot(r, print.auc=TRUE)